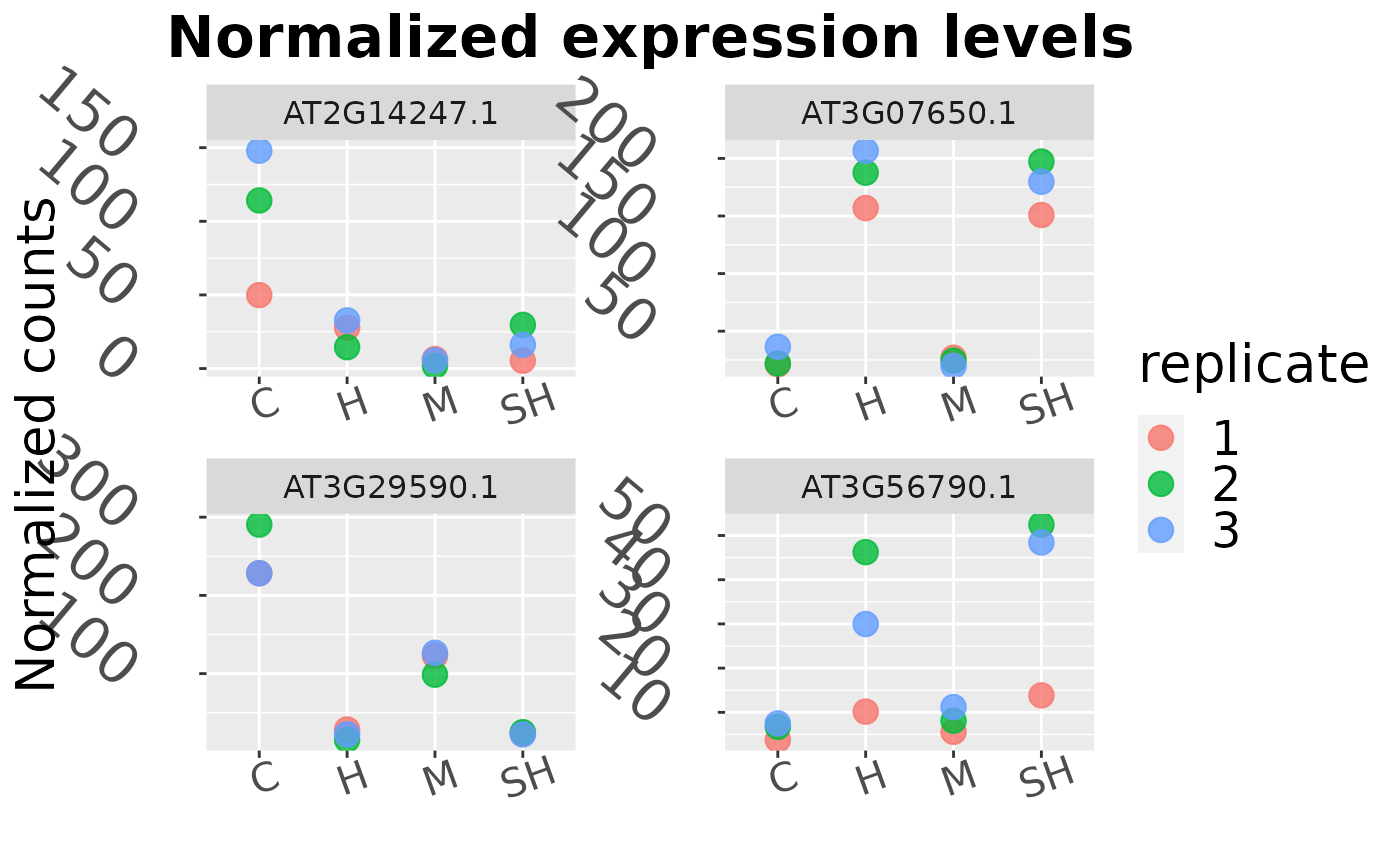
The normalized counts of the desired genes in the specified conditions are shown. Please limit the number of input genes for readability reasons (up to 10 genes).
draw_expression_levels(
data,
genes,
conds = unique(stringr::str_split_fixed(colnames(data), "_", 2)[, 1]),
gene.name.size = 12,
log2_count = FALSE,
start_from_zero = FALSE
)Arguments
- data
normalized expression dataframe, with genes as rownames and conditions as colnames.
- genes
character vector of genes to be plotted (must be contained in the rownames of data)
- conds
conditions to be shown on expression levels (must be contained in the column names of data before the _rep suffix). Default : all conditions.
- gene.name.size
size of the facet plot title font for each gene. Default : 12
- log2_count
transform count using the log2 function. A pseudocount of 1 is added to avoid negative values.
- start_from_zero
set the beginning of the y axis to 0.